Advancing sequencing and computational methods to identify genetic and phenotypic variations in Staphylococcus aureus from patients on mechanical ventilation
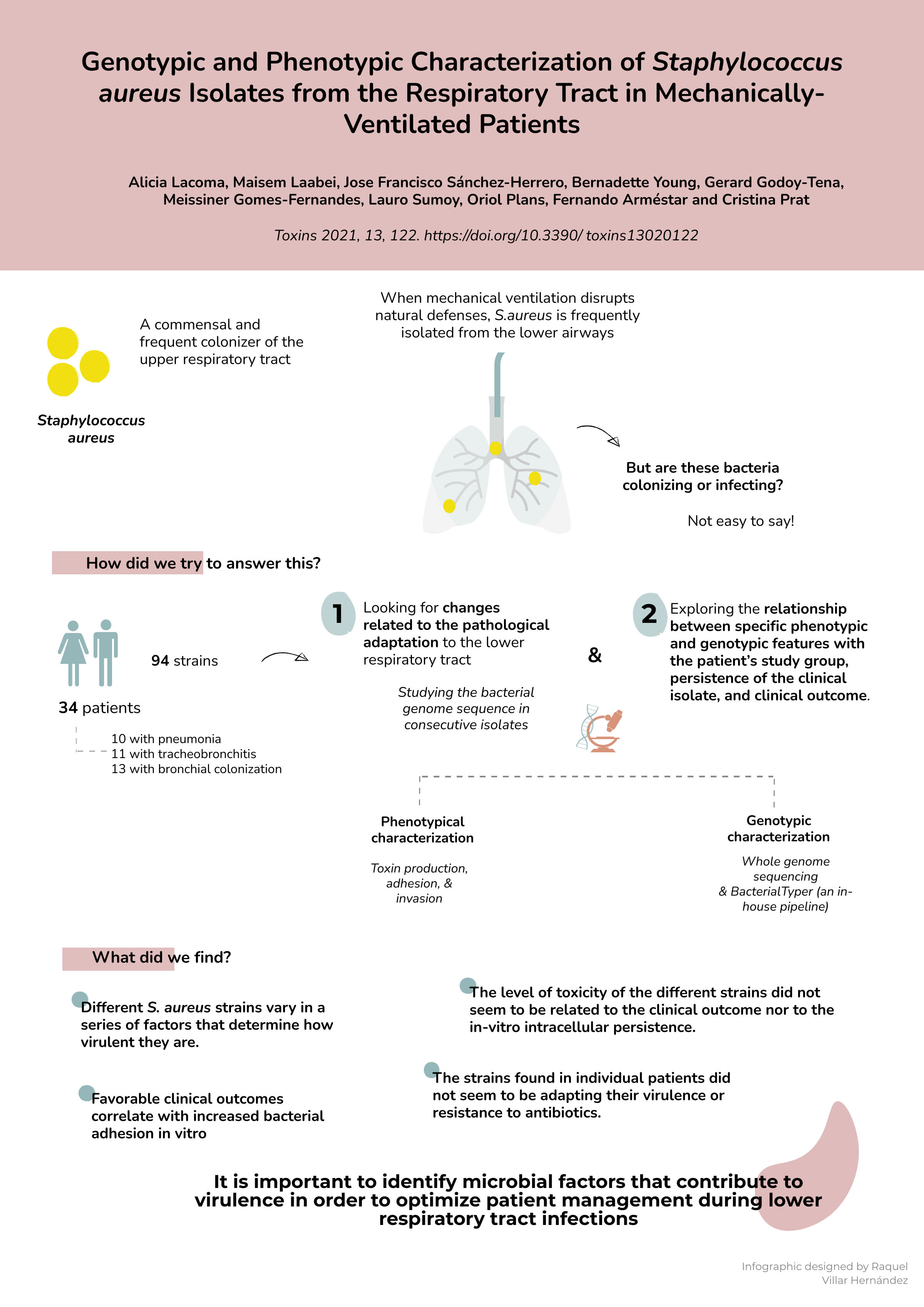
Staphylococcus aureus is a common microorganism that causes different effects ranging from a complete lack of symptoms to organ failure, septicaemia and death. Its presence is not necessarily a sign of infection. S. aureus is often found colonizing the upper respiratory tract or the lungs harmlessly, but in some people an infection develops and patients on ventilators are at a high risk of developing life-threatening infections and complications. This poses a problem for doctors, since it is not easy at early stage to distinguish if S. aureus found in patients is benign, or the sign of serious illness. There is also a suggestion that the microbes can adapt and change within patients to become more virulent and start causing infection.
"Our study was set up to look at the characteristics and the genetic make-up of the different strains of S. aureus that were obtained from samples from patients on mechanical ventilation," explains Cristina Prat of the Innovation in Respiratory Infections and Tuberculosis Diagnosis Group at the Germans Trias i Pujol Research Institute (IGTP) and the CIBER Network. "We looked to see if the genome of the microbes was changing as a result of selection to adapt to their environment in the lungs and to compare the characteristics of the bacteria with the clinical outcomes to provide more information for doctors," she added. The research has been published in the journal Toxins.
The group analysed a set of 94 strains, isolated over time from 34 patients. They performed a complete characterization of their physical features and sequenced their genomes using sophisticated computational methods to identify the relationships between the strains. A bioinformatics pipeline was developed for this purpose at the High Content Genomics and Bioinformatics Unit at IGTP, led by Lauro Sumoy. The tool, named Bacterial Typer, has been implemented by Jose Francisco Sanchez Herrero. While it was tuned specifically for S. aureus for this work, it can be used to study of other microorganisms as well.
"Our results do show that the different strains of S. aureus do vary in a series of factors that determine how virulent they are," explains Alicia Lacoma, first author on the paper. However, the researchers did not find that the strains found in individual patients were adapting their virulence or resistance to antibiotics. Neither did they find that the level of toxicity of the different strains was related to the outcome for the patients. "We did find one difference in strains of the bacteria in patients with pneumonia and poorer outcomes," Lacoma continues. "It is very important to find ways for doctors to decide how to manage patients depending on the type of infection by S. aureus they have. We have now developed these powerful tools to study the bacteria isolated from patients and will continue to look for ways to differentiate between the outcomes they produce."
This work was performed in close collaboration with Fernando Arméstar of the Intensive Care Unit, Hospital Germans Trias i Pujol. The study was made possible by funding from the Instituto de Salud Carlos III, of the Spanish Government, in conjunction with the Regional Development Funds from the European Commission. It also received funding from the CIBER Respiratory Disease Spanish research network.
Original Article
Lacoma, A.; Laabei, M.; Sánchez-Herrero, J.F.; Young, B.; Godoy-Tena, G.; Gomes-Fernandes, M.; Sumoy, L.; Plans, O.; Arméstar, F.; Prat, C. Genotypic and Phenotypic Characterization of Staphylococcus aureus Isolates from the Respiratory Tract in Mechanically-Ventilated Patients. Toxins 2021, 13, 122. https://doi.org/10.3390/toxins13020122
